Diagnosis Tool
Disclaimer
The diagnosis tool is currently under heavy development. The results of the tool MUST NOT be used as a clinical diagnosis. We will push periodic updates to improve the tool and add new functionalities. Bugs are expected, if you find any please write a mail to [email protected] indicating the issue. Feedback is greatly appreciated, if you have any suggestions or comments please feel free to share them with us.
The tool is provided "as is", without warranty of any kind, express or implied. In no event shall the authors or copyright holders be liable for any claim, damages or other liability arising from, out of or in connection with the tool or the use or other dealings in the tool.
What is the Diagnosis Tool?
The diagnosis tool allows using the diagnosis Machine Learning model to obtain the probability of multiple diagnoses. It offers an interactive user interface to score the degree of fat infiltration in the muscles and allows to export or import the data for future use.
You can access the diagnosis tool here.
Getting Started
The Diagnosis Canvas
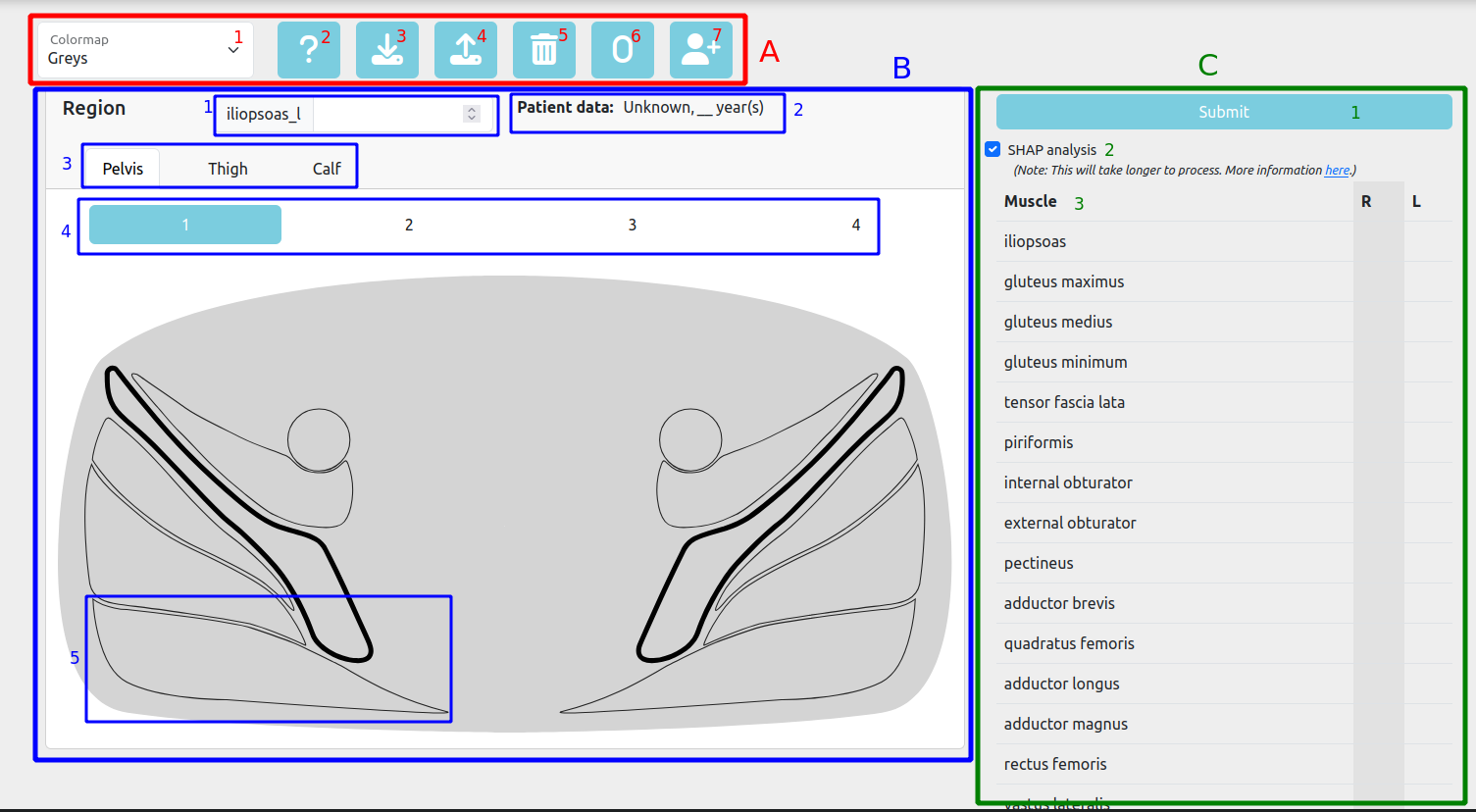
- Toolbar
- Colormap
- Help
- Download scores
- Upload scores
- Clear scores
- Set to zero
- Add patient info
- Muscle Diagrams
- Selected muscle score
- Patient info
- Region selector
- Diagram selector
- Muscle
- Mercuri Table
- Submit button
- SHAP analysis checkbox
- Scores table
Toolbar
Colormap
The Colormap dropdown allows you to select the colormap used to represent the Mercuri scores in the Mercuri table and the regions panel. The default colormap is "Greys", but you can choose between multiple options.
Help
This redirects to this section of the documentation.
Download scores
The Download scores button allows you to download the current Mercuri scores in CSV format. The downloaded file will have the same structure as the one used to upload scores.
Upload scores
The Upload scores button allows you to upload a CSV file with the Mercuri scores. The uploaded file must have the same structure as the one downloaded using the Download scores button. The uploaded scores (as well as the age and sex) will replace the current ones.
Clear scores
The Clear scores button allows you to clear the current Mercuri scores. This will set all the scores to "missing". The age and sex will remain unchanged.
Set to zero
The Set to zero button allows you to set all the Mercuri scores to 0.
Add patient info
The Add patient info button opens a modal to enter the age and sex of the patient. The age can be an integer or have decimals. If the entered age is outside of the age range seen by the model during training the model could produce unexpected results. A warning message will appear to indicate this.
Muscle Diagrams
Selected muscle score
This shows the current Mercuri score of the selected muscle. The score can be empty (blank) or have a value between 0 and 4. You can modify the score by clicking on each muscle and entering a value here, or scrolling over the muscles.
Patient info
The Patient info shows the age and sex of the patient. You can modify these using the "Add patient info" button.
Region selector
The Region selector allows you to select the region of the leg you want to score. The available regions are: Pelvis, Thigh and Calf. Each region has its own set of diagrams.
Diagram selector
The Diagram selector allows you to select the diagram you want to display. Each region has its own set of diagrams. The diagrams are axial MRI slices of the leg, showing the muscles and their current Mercuri scores (colour-coded).
Muscle
The diagrams are composed by multiple interactive muscles. The muscles are colour-coded according to their current Mercuri score. You can interact with the muscles to increase or decrease their Mercuri score by either scrolling over the muscle or clicking it and modifying the score in the input box. If hovered, the muscle name will be displayed in a tooltip.
Mercuri Table
Submit button
The Submit button allows you to submit the current Mercuri scores to the machine learning model. Once you click it, a loading screen will show while the model is predicting the diagnostic probabilities. Once this is done, you will be redirected to the results page.
SHAP analysis checkbox
The SHAP analysis checkbox allows you to enable or disable the SHAP analysis. If enabled, the SHAP values will be calculated and displayed in the results page. This is a computationally expensive operation and can take a while to complete. More information about SHAP analysis can be found here
Scores table
The Scores table shows the current Mercuri scores for the muscles, distinguishing between the right and left sides. The table is divided into two sections: the left section shows the right side muscles and the right section shows the left side muscles. The muscles are colour-coded according to their current Mercuri score. You can interact with the muscles to increase or decrease their Mercuri score by either scrolling over the muscle or clicking it and modifying the score in the input box.
The Diagnosis Result

- Download results
- First choice of the model
- Pie chart of the estimated probabilities
- Results table
- SHAP analysis results
Download results
The Download results button allows you to download the current results in CSV or PDF format.
First choice of the model
This is the diagnosis with the highest probability, as predicted by the model. The model can give multiple diagnoses with high probabilities.
Pie chart of the estimated probabilities
The pie chart shows the estimated probabilities for each diagnosis. The pie chart is interactive and you can hover over each slice to see the exact probability.
Results table
The results table shows the estimated probabilities for each diagnosis. The table is sorted by probability, from highest to lowest. The table can be sorted with different methods using the dropdowns.
SHAP analysis results
The "SHAP" column show a waterfall plot with the SHAP values for each diagnosis. The plot can be enlarged by clicking on it. These plots are useful for understanding how the model arrived at its decision and validating the model logic.
Mercuri Score
The Mercuri Score is a semi-quantitative scale to measure the fat replacement in muscles. To learn more about the Mercuri score, whatch the video below.
Data Imputation
Missing data is uncompatible with many Machine Learning methods, including the ones used by MYO-Guide. Ideally, the MRIs should cover all the leg volume, from the Psoas to the Flexor Hallucis/Digitorum Longus muscles. Age and sex should also be available. However, missing data is unavoidable when the MRI does not cover the full volume of the leg or any image artefact making scoring a muscle impossible. For these cases, MYO-Guide implements a data imputation mechanism that attempts to approximate the missing values.
While this mechanism introduces some bias, it is useful when some muscles are missing. However, if a whole region is missing or there is a large proportion of missing data, the results of the tool should be taken carefully. If the SHAP analysis gives high SHAP values to missing features, this indicates that the model is relying on the imputed values to make a decision. In this case, the results should be taken with caution.
Supported Diagnostics
The diagnosis tool currently supports the following diagnoses:
| Gene | Disease | Other Names |
|---|---|---|
| ANO5 | Anoctaminopathy | LGMD-R12, ANO5 muscle disease, LGMD2L |
| CAPN3 | Calpainopathy | LMGD-R1, LGMD-D4, LGMD1I, LGMD2A |
| DMD | Dystrophynopathy | DMD, BMD, Duchenne / Becker muscular dystrophy |
| FKRP | FKRP related LGMD / Congenital muscle dystrophy | LGMD-R9, LGMD2I, MDC1C |
| DMPK | Myotonic dystrophy type I | MD1, DM1, Steinert disease, dystrophia myotonica type 1 |
| SMN-1 | Spinal muscular atrophy | SMA, Werdnig Hoffmann disease, Kugelberg Welander disease |
| GNE | GNE myopathy | Nonaka disease, hereditary inclusion body miositis (HIBM), IBM2, distal myopathy with rimmed vacuoles |
| GAA | Pompe Disease | Glycogen storage disease type II, acid alpha-glucosidase deficiency, acid maltase deficiency |
| LMNA | Laminin alpha 2 muscular dystrophy | LGMD1B |
| PABPN1 | Oculopharyngeal muscle dystrophy | PABP2, OPMD |
| DYSF | Dysferlinopathy | LGMD-R2, Miyoshi myopathy, distal anterior compartment myopathy, LGMD2B |
| DUX4 | Facio-Scapulo-Humeral muscular dystrophy type 1 | D4Z4, FSHD1 |
| SarcoG | Sarcoglycanopathy (Alfa/Beta/Gamma) | LGMD-R3, LGMD2D / LGMD-R4, LGMD2E / LGMD-R5, LGMD2C |
| TTN | Titinopathy | LGMD-R10, LGMD2J |
| VCP | Vallosin containing protein (VCP) myopathy | Inclusion body myopathy associated with Paget's disease of bone and frontotemporal dementia (IBMPFD) |
| OPDM | Oculopharyngodistal myopathy | |
| HypoPP | Hypokalemic periodic paralysis | |
| PYGM | Glycogen storage disease type V | McArdle disease |
| CLCN1 | Myotonia congenita | Chloride channel myotonia |
| SCN4A | Hyperkalemic periodic paralysis | Sodium channel myotonia |
SHAP Explainability Analysis
SHAP (SHapley Additive exPlanations) is a unified framework for interpreting predictions made by machine learning models. It is based on the concept of Shapley values from cooperative game theory, which were originally developed to fairly allocate payouts among players in a coalition. SHAP adapts this concept to the context of machine learning, attributing the contribution of each feature to a specific prediction in a principled and consistent manner. SHAP values explain individual predictions, making it easier to analyse specific cases. By analysing the individual predictions of all samples used for training, we are able to obtain global insights about the model decision-making process and, therefore, about the underlying patterns of muscle involvement useful for diagnosis.
SHAP values
SHAP values quantify the contribution of each feature to the difference between the model's output for a specific sample and the mean model output across the dataset. Formally, the SHAP value for a feature is calculated as the weighted average of its marginal contributions across all possible coalitions of features. A SHAP value is calculated for each feature of each sample for each target diagnosis; indicating the impact each feature has on the predicted probability of each diagnosis, for each sample. In one of our studies, we trained a model with 2961 samples, 37 features and 20 possible diagnoses; resulting in a [2961x37x20] matrix of SHAP values. Positive SHAP values indicate an increase on the model output (predicted probability of the given disease), while negative values indicate a decrease. A value close to 0 indicates low impact on the model output. For example, if a sample's vastus medialis SHAP value for GNE is high, this indicates that the amount of intramuscular fat of the vastus medialis (whether high or low) for this sample in specific increases the predicted probability of GNE.
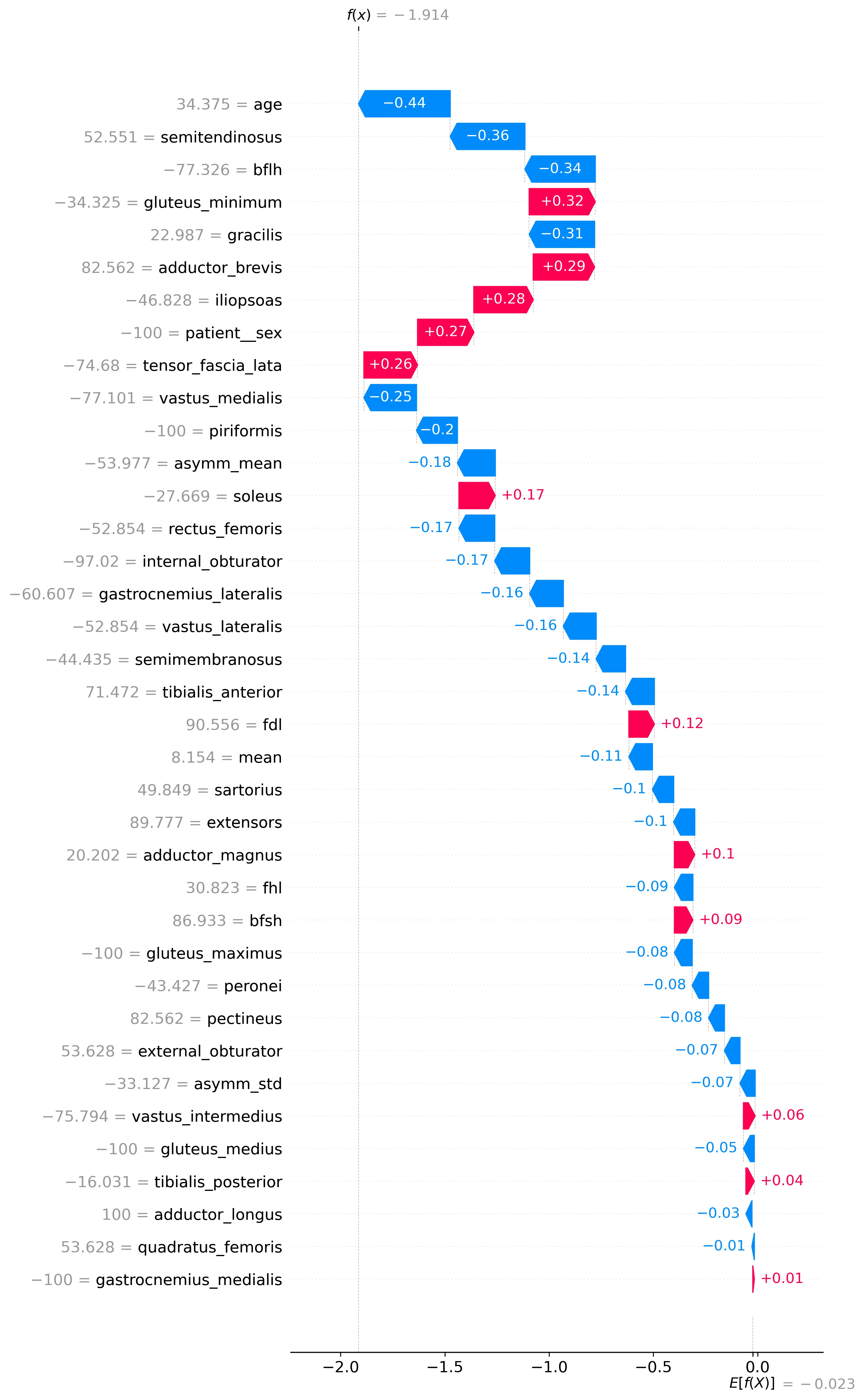
SHAP values Waterfall Plot
Waterfall plots help interpret how Myo-Guide arrives at a specific prediction for an individual case, breaking down the prediction process step by step. The plot starts with the base value, which is typically the average model output across the training data. From there, each bar represents the SHAP value of a feature—indicating how much that feature has pushed the model's prediction up or down relative to the base. Positive SHAP values (which push the prediction higher) and negative SHAP values (which push it lower) are visualized as bars moving left or right, respectively. By stacking these contributions in order, the waterfall plot shows exactly how the model combines the effects of individual muscle fat features to reach its final diagnostic prediction. This makes it easier to understand and trust the model's decision, especially in complex or critical cases. Features are sorted by importance, with the most impactful at the top and the least at the bottom. The plot also show the value of each feature, please note that all these values are normalised to a range of -100 to 100 (including age, sex, asymmetry, ...). Note that the model output (x axis) represents the raw logits of the model, not the a probability between 0 and 1.
Limit of Diagnosis Requests
The number of requests each user can have in the dashboard is currently limited to 30. This has been done to keep under control the memory consumption in the database and ensure the operanting costs of the platform are kept low, allowing us to maintain Myo-Guide in the long-term. The Myo-Guide team plans to re-evaluate this limit in the future based on overall user usage and the costing of our hosting services. If you reach your limit and wish to create additional requests, you can remove some requests to create some space. The results of a request can be downloaded and kept separately if needed using the download option in the results page.